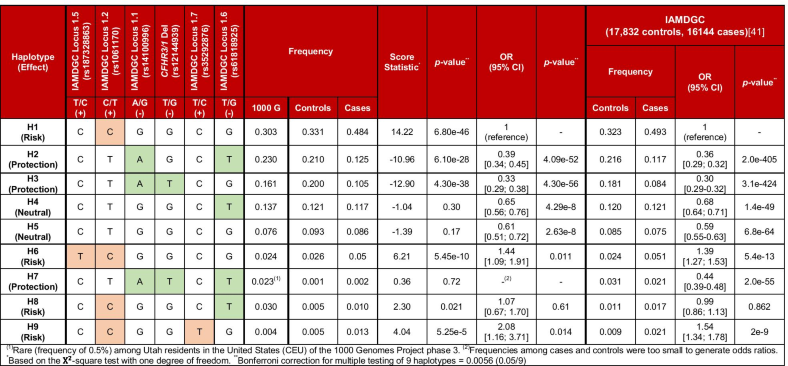
- The labelling and numbering of haplotypes follows that of the haplotype analysis of the IAMDGC cohort [41]. For consistency with this analysis, the rare variant rs35292876 (IAMDGC Locus # 1.7 with minor allele T, MAFcontrols = 0.005; MAFcases = 0.014; OR 2.99 [1.73; 5.17], p = 8.8e−5 in our cohort and MAFcontrols = 0.009; MAFcases: 0.021; OR 2.42, p = 8.2e−37 in the IAMDGC study) was also included. The rs35292876 minor allele exists exclusively on a low-frequency haplotype containing a C (risk) allele at rs1061170. While this variant may modulate risk, its frequency and effect size are therefore accounted for by haplotypes with a C allele at rs1061170. Haplotypes in the IAMDGC cohort used rs570618 in place of rs1061170 (r2 = 0.9914, Dʹ = 1.0), rs10922109 in place of rs14100996 (r2 = 0.9919, Dʹ = 0.9959) and rs6677604 in place of rs12144939
- (1)Rare (frequency of 0.5%) among Utah residents in the USA (CEU) of the 1000 Genomes Project phase 3. (2)Frequencies among cases and controls were too small to generate odds ratios *Based on the \({\mathrm{\rm X}}^{2}\)-square test with one degree of freedom. **Bonferroni correction for multiple testing of 9 haplotypes = 0.0056 (0.05/9)

